Credit: University of Oxford
A research team from the University of Oxford has developed a molecular hopper that is small enough to be able to move single strands of DNA through a protein nanotube.
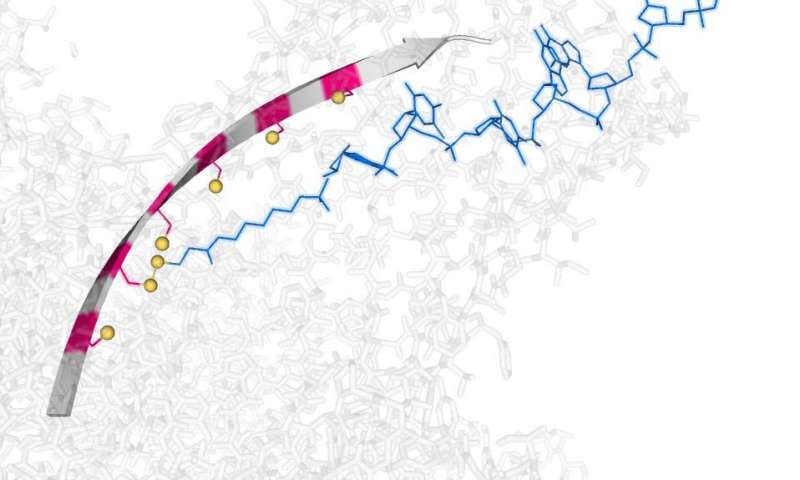
The device works by making and breaking in sequence simple chemical bonds that attach it to a nanoscale track that can be turned on, off or reversed by a small electrical potential.
“Being able to control molecular motion is the holy grail of building nanoscale machines,” professor Hagan Bayley of Oxford University’s Department of Chemistry said in a statement. “Being able to process single molecules of DNA under precise chemical control may provide an alternative to the use of enzymes in DNA sequencing technologies, improving their speed and the number of molecules that can be analyzed in parallel.”
The hopping motion is based on three sulfur atoms, which occur in water at room temperature. The hopper, which is powered and controlled by an electric field, then takes sub-nanometer steps. Scientists can control the direction of the hoping by reversing the electric field.
A ratcheting motion is required for nanopore sequencing, which at present is achieved by using an enzyme. In the new device, the hopping motion is a chemical ratchet, which could be applied to DNA and RNA sequencing due to the step-size being similar to the inter-nucleotide distance in single-stranded DNA.
Previously, the researchers were able to construct molecules with sliding and rotating elements, technology that resulted in the 2016 Nobel Prize. Since then, the researchers discovered a way to produce molecules that make sub-nanometer hopping steps that can be detected one at a time and are subject to external control.
Each step takes approximately a few seconds for the hopper to complete and the team is hoping to increase the speed of the chemistry as well as the length of the track that is currently limited to six footholds.
The study was published in Science.
Filed Under: Genomics/Proteomics



