The enzyme encoded by a single gene, cytochrome P450-2D6 (CYP2D6), is responsible for metabolizing about 25 percent of all commonly used drugs, making it one of the most pharmacologically important genes in the human genome. Variants within this gene can help predict how patients respond to medications ranging from painkillers to antipsychotics, which makes CYP2D6 an essential gene to consider when implementing pharmacogenomics into clinical care. In addition, a thorough picture of the landscape of CYP2D6 genetic variability could also benefit drug development research by helping understand the variable metabolism of new drugs and informing dosing decisions during clinical trials.
However, CYP2D6 is not a typical gene. There are more than 100 known allelic variations of CYP2D6, including deletions and duplications, and other structural rearrangements — and its complex sequence is made even more challenging to interrogate by a highly homologous nearby pseudogene. Consequently, the currently available FDA-approved molecular diagnostic assays only genotype a small fraction of known alleles, and research methods based on Sanger or short-read sequencing typically cannot adequately resolve the genetic complexity of the CYP2D6 locus.
Given that a comprehensive understanding of CYP2D6 variation is essential for any research on medications metabolized by CYP2D6, we and other scientists have recently adopted long-read single-molecule real-time (SMRT) sequencing to better resolve this challenging region. Importantly, subjecting large amplicons that encompassed the CYP2D6 gene to SMRT sequencing resulted in full-length characterization of CYP2D6, including the identification of structural rearrangements and the specific sequence of duplicated alleles.
This approach is now enabling scientists to discover novel CYP2D6 genetic variants, promising a far more exhaustive catalog of CYP2D6 variation in different populations, which could lead to better metabolizer phenotype prediction, and ultimately help identify the right drug at the right dose to the right patient using pharmacogenetics.
The CYP2D6 challenge
First identified in the 1980s, the CYP2D6 enzyme has long been recognized for its role in hepatic drug metabolism. It plays a direct role in both activating some drugs (for example, it metabolizes codeine into morphine) and in deactivating or excreting other drugs.
The CYP2D6 gene lies on chromosome 22, just 10 kb from its nearest pseudogene CYP2D7P1, which is 97percent identical to the CYP2D6 sequence. The ability to sequence CYP2D6 and determine an individual’s star (*) allele ‘diplotype’ is important for scientists in drug discovery, pharmacology, clinical laboratories, and basic research. Commonly used methods to interrogate CYP2D6 include targeted genotyping, which typically assays a small subset of known alleles, and Sanger sequencing, which is tedious and expensive. Short-read sequencing, with reads just a few hundred bases long, has been hindered by ambiguous alignments to the gene or nearby pseudogene.
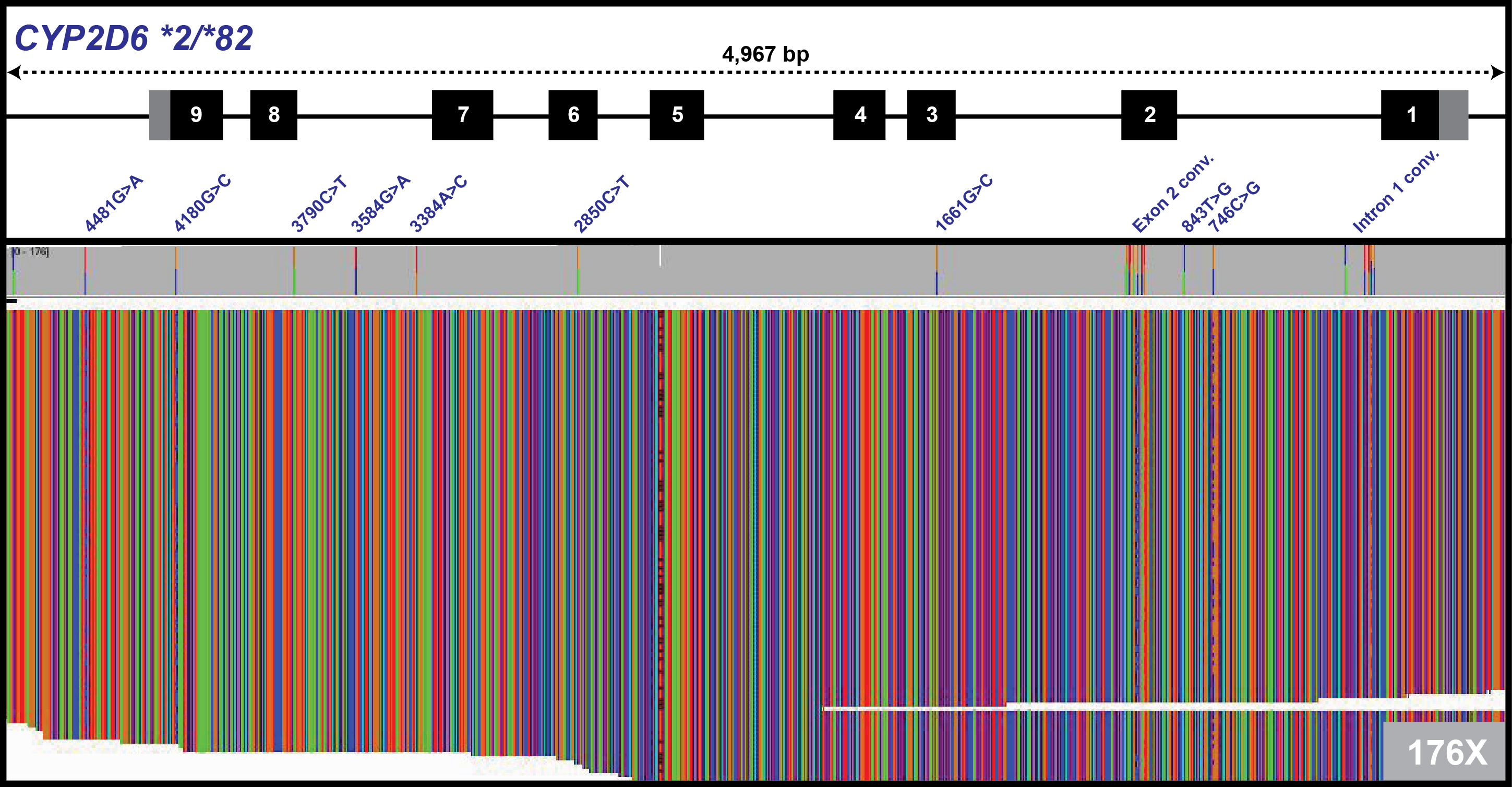
Illustration of long-read single molecule real-time (SMRT) sequencing (Pacific Biosciences) of the CYP2D6 gene locus in one subject with a star (*) allele diplotype of *2/*82. The CYP2D6 gene, which is encoded on the negative strand, is illustrated at the top of the image with exons denoted as numbered black boxes. The identified single nucleotide variants in this subject are noted below the CYP2D6 gene illustration based on their common sequence coordinates used by the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Below the CYP2D6 variants are individual SMRT sequencing reads at a depth of 176X (green = Adenine, orange = Guanine, blue = Cytosine, red = Thymine). Image courtesy of Drs. Stuart A. Scott and Yao Yang of the Icahn School of Medicine at Mount Sinai, New York.
CYP2D6 is notoriously polymorphic, which includes numerous single nucleotide variants throughout the coding and non-coding regions, structural variants and copy number variation, tandem events, and partial gene conversions. Phasing identified variants into distinct haplotypes is also important, but very challenging. For example, when variant CYP2D6 alleles and a gene duplication are identified, it is essential to determine which allele is duplicated, as some duplicated alleles have increased function whereas others have no function depending on the sequence. However, most currently used technologies cannot clarify which allele is duplicated and only report the existence or absence of a CYP2D6 gene duplication.
Long-read sequencing
Recently, we evaluated the use of long-read sequencing to fully interrogate the entire 5 kb CYP2D6 region. In our study recently published in Human Mutation, we reported the development and assessment of the full gene CYP2D6 SMRT sequencing method using the Pacific Biosciences platform.
Long amplicons were optimized that covered CYP2D6 and its upstream duplicated copy when present, which were barcoded, purified and pooled, and subjected to SMRT sequencing. Using a novel error-correction algorithm to make the most of the long-read data, variant calling was accomplished using a custom GATK-based pipeline that included an amplicon long-read error correction (ALEC) script. The CYP2D6 SMRT sequencing assay validation included three sets of samples: positive controls with previously determined genotype and diplotype data available; copy number variation controls to test the ability to specifically sequence duplicated alleles; and a set of publicly available samples with discordant or unclear CYP2D6 genotyping results from a previously reported CDC GeT-RM reference material study.
The results showed that long-read CYP2D6 SMRT sequencing accurately detected all CYP2D6 alleles and was capable of characterizing duplicated alleles. Even with the challenging discordant or unclear samples, we found that long-read sequencing could clarify their diplotypes.
Although the method development study was not designed for discovery, three novel CYP2D6 alleles were discovered during the assay validation that were not previously reported by the Human Cytochrome P450 (CYP) Allele Nomenclature Committee. In fact, ~20 percent of the samples used to evaluate CYP2D6 SMRT sequencing were revised to either a non-genotyped or novel star (*) allele, which highlights how long-read sequencing can reveal previously unrecognized variation in well-studied genes and specimens that were previously tested by other technologies.
Opportunities for discovery
The ability to fully interrogate the CYP2D6 locus will expand our knowledge of this region’s genetic diversity and facilitate the discovery of novel alleles in different populations, particularly among understudied racial and ethnic groups. However, although CYP2D6 SMRT sequencing will likely result in the identification of more precise diplotypes, increased discoveries of rare and novel star (*) alleles means that functional studies are going to be needed to determine the effect of these low-frequency sequence variants on enzyme activity.
One of the challenges of pharmacogenomics has been the inconsistent results reported between research studies due to different alleles being genotyped and differences in patient populations. By using long-read CYP2D6 SMRT sequencing in diverse clinical trial populations, the community could have far better resources to more accurately predict response of CYP2D6-metabolized drugs across different ethnicities, from the early phases of drug discovery to the physician ultimately issuing a prescription of that medication. This advance, and others like it, should help renew the enthusiasm around pharmacogenomics as a sound approach for improving patient care and reducing the cost and risk of adverse drug events.
Filed Under: Genomics/Proteomics



