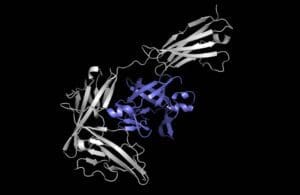
[Image courtesy of the MIT researchers]
The technique represents an improvement on speed by somewhere between 80 and 500 times faster than state-of-the-art software methods and often predicts protein structures that are closer to actual structures that have been observed experimentally when two proteins bind together.
MIT researchers say their method could help scientists gain an understanding into biological processes that involve protein interactions, such as DNA replication and repair, according to the university’s website. It could also potentially increase the pace of new medicine development.
Octavian-Eugen Ganea and Xinyuan Huang are co-lead authors of the paper. MIT co-authors include Regina Barzilay and Tommi Jaakkola.
“Deep learning is very good at capturing interactions between different proteins that are otherwise difficult for chemists or biologists to write experimentally,” Ganea, a postdoc in the MIT Computer Science and Artificial Intelligence Laboratory (CSAIL), said in the website report. “Some of these interactions are very complicated, and people haven’t found good ways to express them. This deep-learning model can learn these types of interactions from data.”
The researchers call their model “Equidock” as it focuses on rigid body docking, the process during which two proteins attach by rotating or translating in 3D space but their shapes don’t squeeze or bend. Equidock takes the 3D structures of two proteins and converts the structures into 3D graphs that the neural network can process, with each amino acid (from which proteins are formed) represented by a node in the graph.
Geometric knowledge was included in the machine learning model to allow for understanding of how objects change when rotated or translated in 3D space. With this, along with mathematical knowledge of how proteins dock in the human body, the system identifies atoms of the two proteins most likely to interact and form chemical reactions, using the points to place the proteins together into a complex.
The researchers incorporated geometric knowledge into the model, so it understands how objects can change if they are rotated or translated in 3D space. The model also has mathematical knowledge built in that ensures the proteins always attach in the same way, no matter where they exist in 3D space. This is how proteins dock in the human body.
Equidock can predict the final protein complex in one-to-five seconds, with baselines taking between 10 minutes to an hour or more. It was often comparable with baselines, but did sometimes underperform in comparison, MIT said.
“We are still lagging behind one of the baselines. Our method can still be improved, and it can still be useful. It could be used in a very large virtual screening where we want to understand how thousands of proteins can interact and form complexes. Our method could be used to generate an initial set of candidates very fast, and then these could be fine-tuned with some of the more accurate, but slower, traditional methods,” he says.
The researchers intend to incorporate specific atomic interactions so Equidock can become more accurate. The technique could also be used to develop small, drug-like molecules to shorten the drug development timeline. Future enhancements include flexible protein docking, for which the researchers are working on synthetic data to use to improve the model.
Research was funded in part by the Machine Learning for Pharmaceutical Discovery and Synthesis Consortium, the Swiss National Science Foundation, the Abdul Latif Jameel Clinic for Machine Learning in Health, the DTRA Discovery of Medical Countermeasures Against New and Emerging (DOMANE) threats program, and the DARPA Accelerated Molecular Discovery program.
Filed Under: Drug Discovery, Drug Discovery and Development



