
Since its invention, next-generation sequencing (NGS) technology has shown promise in many research areas, and recently, the technology has found its way into the clinic, playing an integral role in liquid biopsies for cancer and other diseases. Pathologists can use NGS to detect mutations in cell-free DNA (cfDNA)—the DNA fragments that tumors and other cells shed into the blood—enabling researchers to develop non-invasive genetic screens for a host of conditions.
Despite its promise, however, NGS cannot be effective, and diagnostics cannot be accurate, if the cfDNA library a pathologist creates from a patient sample for sequencing is of poor quality (i.e. contains sheared or degraded DNA) or of incorrect quantity. One method that determines whether a cfDNA library is suitable for sequencing via NGS is automated capillary electrophoresis (CE). CE quantifies and qualifies nucleic acids quickly and helps researchers distinguish low-quality DNA from DNA fit for sequencing. By optimizing cfDNA libraries, CE streamlines and simplifies the NGS workflow and cfDNA analysis, which allows for the development of high volume, noninvasive diagnostics for a variety of diseases.
The Need for Quality Control
Before cfDNA can be sequenced, researchers must first prepare a DNA library, a collection of similarly sized DNA fragments. Several potential problems can arise during library preparation that adversely affect NGS results. If the fragments are low in quality or are of an improper concentration or size, they will yield a suboptimal library, and in turn, yield substandard sequencing results. This concern is apparent in clinics, where blood is often handled by several individuals and stored for a period time before it is tested, which can cause the cfDNA to degrade.
Adding too little or too much library can also lead to difficulties, especially when dealing with clinical samples. Excess library can impede proper sequencing while an insufficient amount of library can hinder the detection of rare targets, resulting in misdiagnosis. Thus, screening a cfDNA library for quality and quantity prior to sequencing is critical, and can ensure that the run will yield high quality data and actionable diagnostic information.
How CE Improves Library Quality
After a researcher isolates cfDNA from a blood sample, CE can be used to size the isolated nucleic acids by running them through a capillary tube, past a fluorescence detector. Besides size, this simple technique also provides valuable information about isolated nucleic acid quality.
Incorporating CE into the NGS workflow for cfDNA addresses several common problems researchers face in building quality DNA libraries. First, while cfDNA comes in various lengths, researchers prefer to study mononucleosomal cfDNA fragments, or fragments created as genomic DNA is sheared around a single nucleosome. CE enables research to measure cfDNA size so the proper fragments can be isolated. Later, researchers add known adapters to 5’ and 3’ ends of fragmented DNA, and CE helps them achieve proper adapter stoichiometry. If a researcher introduces too much adapter during library preparation and they’re not removed properly during clean-up, the adapters can ligate in series, bind to the flow cell, and disrupt a sequencing run. CE can detect samples where the adapter stoichiometry is unbalanced so a researcher can remove them prior to sequencing. Finally, after the cfDNA library is constructed, the fragments’ sizes and the library concentration must fall within a specific range, depending on the NGS method used. In one run, CE assesses the quantity and size distribution of the library fragments to ensure they yield quality sequencing results
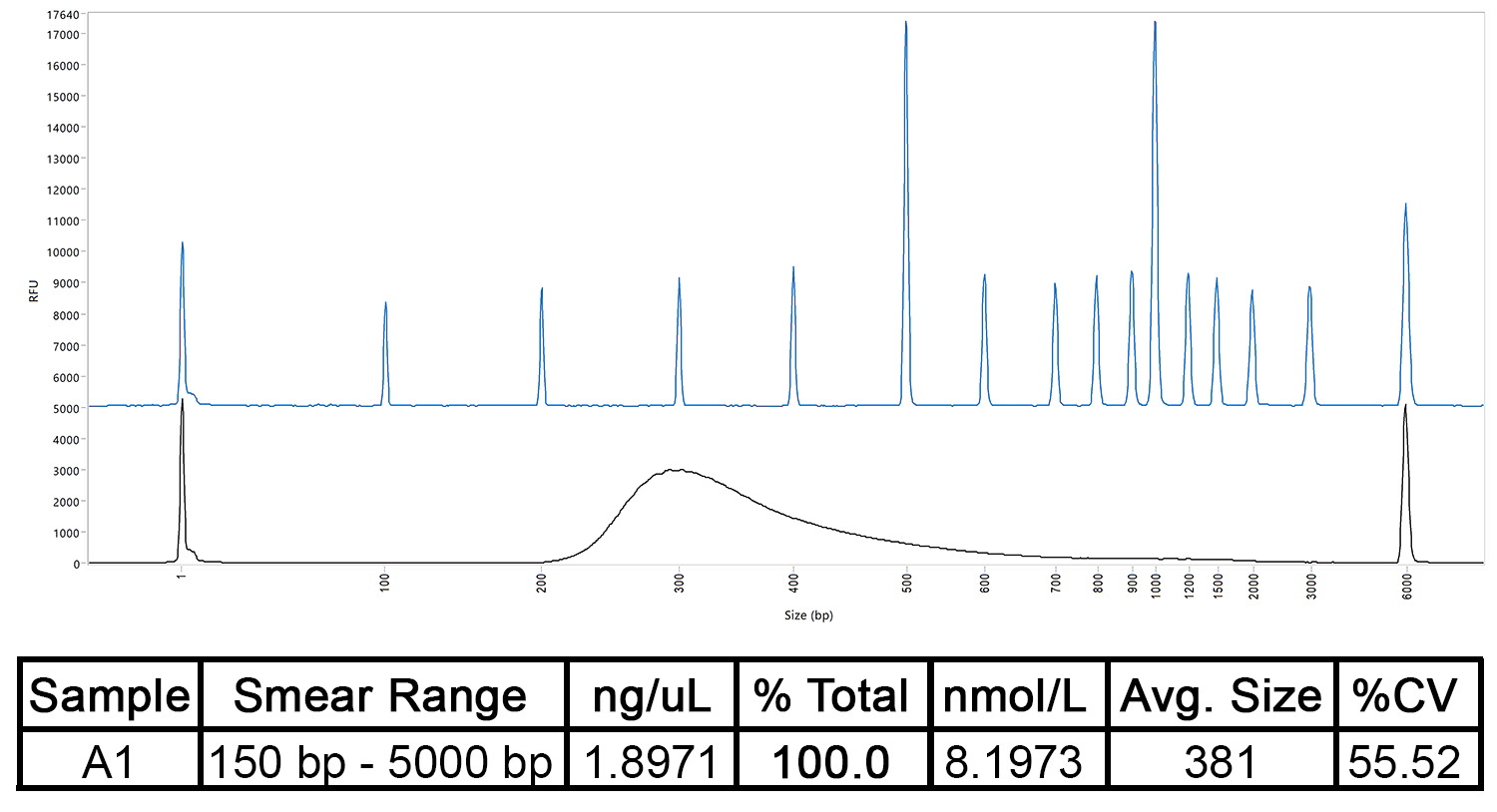
The concentration and size distribution of a DNA library as measured by the Fragment Analyzer Automated CE System, compared to a separation ladder. The table shows the sample’s predicted concentration
Clinical Potential
As an analytical tool that monitors the quality of DNA libraries, CE technology can be applied to virtually any diagnostic that uses cfDNA to detect disease mutations.
Currently, many prenatal genetic screens involve collecting genetic samples from the amniotic sac, a practice that increases the risk of miscarriage. But now, scientists are investigating whether physicians could potentially perform the same tests using the mother’s blood. In fact, researchers have found that the mother’s blood provides genetic clues about fetal health, and consequently, sequencing-based non-invasive prenatal testing (NIPT) is gaining popularity. cfDNA analysis is expected to become central to NIPT soon, which means cfDNA library quality control using CE will become increasingly necessary.
CE can also aid in the analysis of cfDNA samples that might contain cancer biomarkers. As tumor cells recycle, they shed their DNA into the blood, and sequencing these cfDNAs can indicate the presence of cancer before traditional diagnostic techniques, such as imaging, pick up any signs. CE can help clinicians isolate high-quality fragments so sequencing is more likely to yield an actionable diagnosis.
As a simple and accessible biomarker, cfDNA is likely to be used by researchers more widely in diagnostic settings. Researchers who increasingly rely on NGS in prenatal screens and to diagnose conditions such as cancer will need to address the barrier of poor library quality in their preparation. CE is an easy and reliable step they can introduce into their workflows to improve the quality of sequencing-based diagnostics.
Filed Under: Genomics/Proteomics




