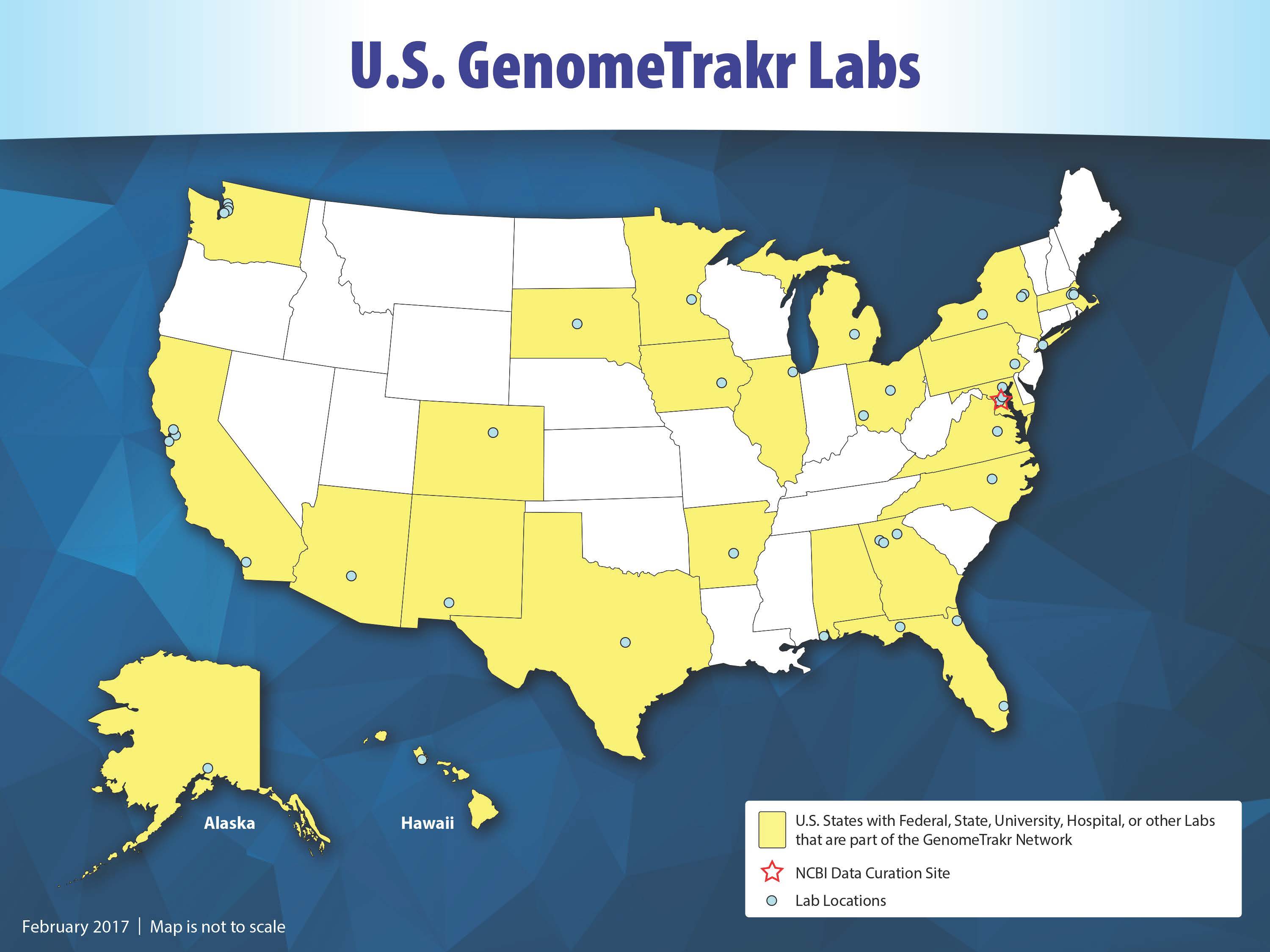
GenomeTrakr consists of 15 federal labs, 25 state health and university labs, 1 U.S. hospital lab, 2 other labs located in the U.S., 20 labs located outside of the U.S., and collaborations with independent academic researchers. Credit: U.S. FDA
The Food and Drug Administration (FDA) is tasked with promoting and protecting public health in many different areas, including ensuring a safe food supply. Over the past 25 years FDA and its sister agencies have worked side-by-side identifying outbreaks (CDC) and tracing back (FDA and USDA) the contamination sources that caused them. Until recently the gold-standard technology for subtyping, or strain identification, of foodborne bacteria has been a molecular technique that reveals a set of banding patterns for each isolate. These patterns are used to identify bacteria and to compare one bacterial isolate to another. However, in the past five years, technology advances have allowed us to rapidly collect data on a foodborne pathogen’s entire genomic sequence, using a process known as whole genome sequencing (WGS). WGS is rapid, precise, automatable, cost-effective, can be universally applied across all foodborne pathogens, and offers higher resolution data for analysis than banding patterns provide. WGS data also allows us to infer the evolutionary history of a set of isolates, which can provide powerful clues for health officials trying to identify the contamination source of an outbreak.
In 2012 FDA began the GenomeTrakr project, a distributed network of state and federal public health labs sequencing microbial foodborne pathogens and uploading the data to a common public database in real-time. The result has been a continuously growing database of genomic sequence information and accompanying metadata (e.g. geographic location, source, and date) from food, environmental, and clinical isolates. Whenever new sequences are submitted they are analyzed and compared to existing sequences in the GenomeTrakr database. Close “matches” between isolates could reveal a foodborne illness outbreak or food contamination event and lead investigators trying to determine how the pathogen got into the food supply.
Paradigm Shift
There are two major paradigm shifts with the GenomeTrakr network. One is the advancement in technology. WGS data provides precise high-resolution information about a pathogen, often enabling us to distinguish which farm or factory a particular pathogen originated from. This discriminatory power, along with full epidemiological concordance, enables us to confidently determine the root cause of more foodborne illness outbreaks than ever before.
The second major paradigm shift is perhaps more significant. GenomeTrakr data is made public immediately after data collection and before the FDA analyzes it. This open data approach has several benefits. One is that anyone can view and analyze the data immediately, without needing to make a FOIA request for the information. Another benefit is that researchers and organizations outside the traditional public health lab networks can contribute to the database. For example, academic researchers can submit sequences from isolates collected through their research efforts. This includes sequences from sources not regularly sampled by traditional public health labs (for example, population-level surveys from the natural environment). An open database also allows for health officials and researchers from around the world to submit and analyze their isolates in a broader global context. This is increasingly important because people now regularly consume ingredients and foods that were sourced from other countries. Having lots of contributors who are submitting sequences for isolates collected from a many different products and environments helps to increase the food and environmental strain diversity in the database and provides a platform for global collaboration of foodborne pathogen surveillance.
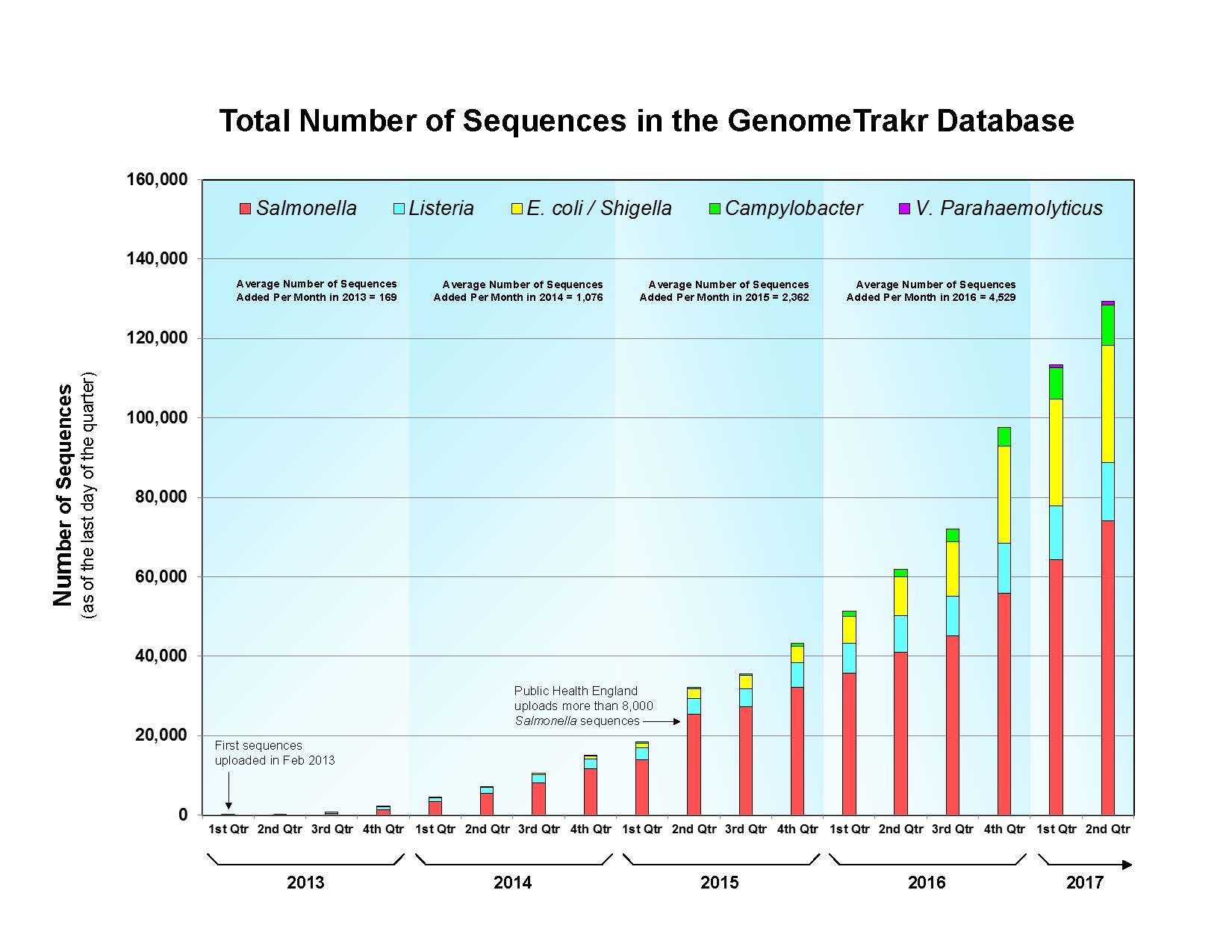
As of August 2017 the five foodborne pathogens under surveillance within GenomeTrakr are: Salmonella enterica, Listeria monocytogenes, Escherichia coli/Shigella, Campylobacter jejuni, and Vibrio parahaemolyticus. Credit: U.S. FDA
GenomeTrakr Data
The raw WGS data and daily analyses are publically available at https://www.ncbi.nlm.nih.gov/pathogens. As of August 2017 the five foodborne pathogens under surveillance within GenomeTrakr are: Salmonella enterica, Listeria monocytogenes, Escherichia coli/Shigella, Campylobacter jejuni, and Vibrio parahaemolyticus.
To date, the Salmonella collection is the largest, with over 80,000 sequences, while the Vibrio parahaemolyticus collection, the newest collection, has about 1,000 sequences. Each morning, FDA scientists mine data from this site, looking at submissions and following up on items of interest. In 2016, the FDA took over 200 regulatory actions based on data derived from GenomeTrakr.
This is how the FDA typically uses the data. A sequence from an isolate taken during a routine FDA inspection and submitted to the database might “match” a set of clinical isolate sequences. This would prompt the FDA to start an inquiry, which might lead to a follow-up inspection and more sampling at the location from which the environmental isolate was originally collected, in an effort to determine if there is causal link between processing facility and the illness. If a link were established, appropriate regulatory action would be taken.
In another example, a non-FDA user of the database might be curious about tracking a current outbreak. This user could search the database for words related to the outbreak (ex. papaya, sprouts, ice cream, or flour) to find clusters (groups of closely related strains), metadata, and raw sequence data associated with the outbreak or event of interest.
GenomeTrakr data can also be monitored for emerging antibiotic resistance (AMR) genes. Along side the cluster results, a panel of AMR genotypes is predicted for each genome, so a user can track the incidence of a particular resistance gene across the database or be alerted when new patterns emerge.
The examples given here are just a few applications of the GenomeTrakr database. GenomeTrakr is a great model of a federally funded program whose initial small investment is returning ten-fold in ways we could never predict at the onset.
Eric Stevens, Marc Allard, and Eric Brown, FDA co-authored this piece.
Filed Under: Genomics/Proteomics




