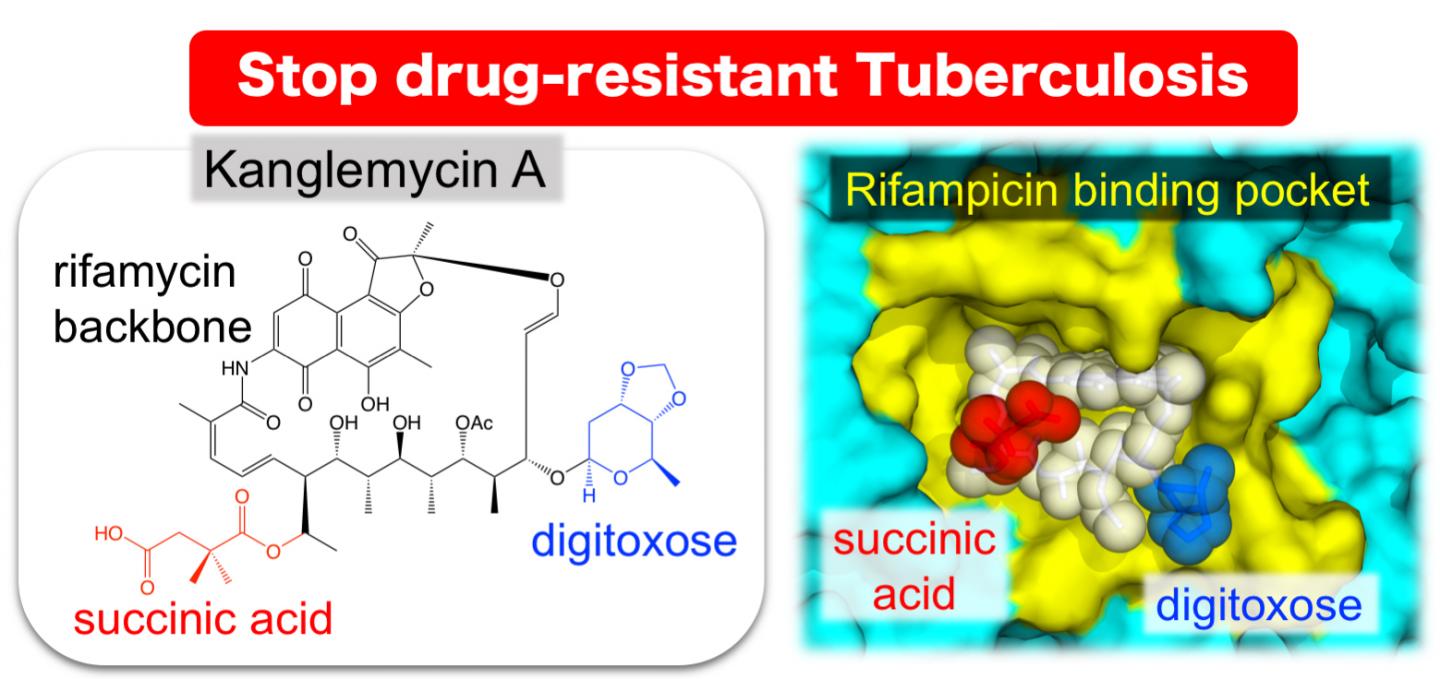
The natural antibiotic kanglemycin A binds bacterial RNA polymerase at the rifampicin binding-pocket, but maintains potency against rifampicin-resistant mutants due to two unique chemical groups (digitoxose and succinic acid) that increase its affinity to rifampicin-resistant RNA polymerase by binding just outside the rifampicin-binding pocket.
A naturally occurring antibiotic called kanglemycin A is effective against Mycobacterium tuberculosis, the bacteria that cause tuberculosis, even in drug-resistant strains, according to an international team of researchers who used chemistry, molecular biology, microbiology, and X-ray crystallography to show how the compound maintains its activity. A paper describing the research appears September 20, 2018 in the journal Molecular Cell.
The compound, kanglemycin A, is related to the antibiotic rifampicin, according to Katsuhiko Murakami, professor of biochemistry and molecular biology at Penn State and one of leaders of the project. “Rifampicin is already part of the cocktail of antibiotics used to treat tuberculosis, but many strains of the tuberculosis-causing bacteria have developed resistance to it,” Murakami said.
“Tuberculosis is the leading cause of death by infectious disease worldwide,” said Murakami. “Development of rifampicin resistance in M. tuberculosis has made treatment of this disease very difficult since it extends treatment time of tuberculosis from 6 months to 2 years. Identifying new compounds that are effective against the rifampicin-resistant bacteria is incredibly important for public health.”
The researchers screened a library of naturally occurring compounds from U.K. biotech company Demuris Ltd. for their ability to inhibit bacterial cell growth or prevent the production of RNA–an essential process in all living organisms–in bacteria. They discovered that a compound named kanglemycin A was effective at inhibiting RNA production even in rifampicin-resistant bacteria.
“Kanglemycin A is related to rifampicin, an antibiotic that functions by binding to bacterial RNA polymerase, the enzyme responsible for RNA production, and preventing it from making more RNA,” said Murakami. “Understanding how kanglemycin A manages to maintain its affinity to rifampicin-resistant RNA polymerase and stay active against the drug-resistant bacteria will help to accelerate its approval for use in patients with tuberculosis.”
To determine the mechanism of kanglemycin A action against rifampicin-resistant RNA polymerase, the Murakami group used X-ray crystallography to determine the three-dimensional structure of the complex of kanglemycin A bound to bacterial RNA polymerase. It was known that rifampicin binds to a groove in the RNA polymerase molecule and that mutations that change the amino-acid sequence of the RNA polymerase can prevent this binding, while maintaining the ability to produce RNA. Kanglemycin A binds to the same groove, but its structure revealed extensions that also bind just outside the groove allowing it to inhibit activity of rifampicin-resistant RNA polymerase.
“The X-ray structure actually revealed that kanglemycin A has two modifications that improve its function compared to rifampicin,” said Murakami. “First, one of modifications allows it to bind just outside of the rifampicin binding pocket increasing the strength of its affinity to the RNA polymerase in rifampicin-resistant bacteria. Second, another modification actually allows kanglemycin A to stop the synthesis of RNA even earlier than rifampicin.”
“It is a really exciting finding,” said Nikolay Zenkin, professor of molecular biology at Newcastle University in the U.K. and one of leaders of the project. “The previously unknown interactions of the unique chemical groups of kanglemycin A with RNA polymerase will direct the development of antibiotics against rifampicin-resistant M. tuberculosis. Approximately one third of the world’s population is already infected with M. tuberculosis, and 600,000 people every year are diagnosed with rifampicin-resistant tuberculosis. Our work is the first step in developing a new drug for the treatment of these patients.”
Filed Under: Infectious Disease




